Practice Exercises - Gene Expression and Regulation - AP Biology Premium 2024
Practice Questions
Multiple-Choice
1. The genome of a newly discovered virus has the following nucleotide composition: 22% guanine, 16% cytosine, 34% adenine, and 28% uracil. Based on the nucleotide composition, the genome of this virus is most likely made of which of the following?
(A) single-stranded DNA
(B) double-stranded DNA
(C) single-stranded RNA
(D) double-stranded RNA
2. Prokaryotic genomes are packaged into _________ chromosomes in the _________.
(A) circular; nucleoid region
(B) circular; nucleus
(C) linear; nucleoid region
(D) linear; nucleus
3. Energy is required to separate the two strands of the DNA double helix because of the hydrogen bonds between the base pairs. Based on the base pair content, which of the following would require the least amount of energy to separate the strands of DNA?
(A) 20% G, 20% C, 30% A, 30% T
(B) 30% G, 30% C, 20% A, 20% T
(C) 15% A, 15% T, 35% G, 35% C
(D) 25% G, 25% C, 25% A, 25% T
4. Which of the following correctly describes a structural difference between DNA and RNA?
(A) DNA has a five-carbon sugar; RNA has a four-carbon sugar.
(B) DNA has thymine; RNA has uracil.
(C) DNA has adenine; RNA has cytosine.
(D) DNA is typically single-stranded; RNA is typically double-stranded.
Questions 5 and 6
N-15, also known as heavy nitrogen, is an isotope of nitrogen that is heavier than the isotope that is typically found in nature, N-14. Conducting chemical reactions in the presence of different isotopes of nitrogen allow a scientist to follow nitrogen atoms in a metabolic pathway. In a classic experiment, Meselson and Stahl allowed parent DNA (containing N-15) to replicate in the presence of N-14.
5. After one round of DNA replication, which of the following results would support the statement “DNA replication is semiconservative”?
(A) All DNA molecules have one strand containing N-14 and one strand containing N-15.
(B) 50% of the DNA molecules only contain N-14, and 50% of the DNA molecules only contain N-15.
(C) All DNA molecules only contain N-15.
(D) All DNA molecules only contain N-14.
6. After two rounds of DNA replication, which of the following results would support the statement “DNA replication is semiconservative”?
(A) All DNA molecules have one strand containing N-14 and one strand containing N-15.
(B) 50% of the DNA molecules only contain N-14, and 50% of the DNA molecules only contain N-15.
(C) All DNA molecules only contain N-15.
(D) 50% of the DNA molecules contain only N-14, and 50% of the DNA molecules have one strand containing only N-15 and one strand containing only N-14.
__________________________________________________________________________
(A) One stand of the double helix contains thymine, and the other strand contains uracil.
(B) Only G and C nucleotides appear on one strand of the double helix, and only A and T nucleotides appear on the other strand of the double helix.
(C) RNA polymerase can only synthesize RNA primers on one strand of the double helix.
(D) DNA polymerase can only add new nucleotides in the 5′ to 3′ direction.
8. The enzyme _________ unwinds the double helix of DNA, and the enzyme _________ relieves the supercoiling created by this unwinding.
(A) helicase; topoisomerase
(B) DNA polymerase; topoisomerase
(C) DNA polymerase; ligase
(D) helicase; ligase
9. The enzyme _________ adds new nucleotides to both the lagging and leading strand, and the enzyme _________ joins the discontinuous segments synthesized on the lagging strand.
(A) helicase; topoisomerase
(B) DNA polymerase; topoisomerase
(C) DNA polymerase; ligase
(D) helicase; ligase
10. Which of the following are small, circular pieces of extranuclear DNA that can be found in either prokaryotes or eukaryotes?
(A) Okazaki fragments
(B) RNA primers
(C) plasmids
(D) linear chromosomes
Short Free-Response
11. Reverse transcriptase is an enzyme that makes a complementary DNA copy of RNA in retroviruses. This DNA copy can then insert itself into the genome of the host cell. Reverse transcriptase has a higher error rate than DNA polymerase, which results in more mutations in the DNA copy of the RNA. Reverse transcriptase is not typically used by eukaryotic cells for any function.
(a) Describe which nucleotides you would expect to find in the genome of a virus that uses reverse transcriptase.
(b) The human immunodeficiency virus (HIV) contains RNA as its genetic material. Reverse transcriptase inhibitors have been shown to be effective in slowing the replication of HIV. Explain why reverse transcriptase inhibitors have few side effects in eukaryotic cells.
(c) Predict the rate of mutation in a retrovirus compared to that of a DNA virus.
(d) Justify your prediction from part (c).
12. An experiment is conducted to study the effect of a ligase inhibitor on DNA replication.
(a) Describe the function of ligase in DNA replication.
(b) Identify an appropriate control for this experiment.
(c) Predict which strand of the DNA would be most affected by a ligase inhibitor.
(d) Justify your prediction from part (c).
Long Free-Response
13. Polymerase chain reaction (PCR) uses a heat-stable DNA polymerase and repeated cycles of DNA replication to amplify specific sequences of DNA. Primers specific to the desired DNA sequences are used to direct DNA polymerase to the beginning of the sequence to be amplified. The following table shows the number of copies of the DNA sequence that exist at the end of each cycle.
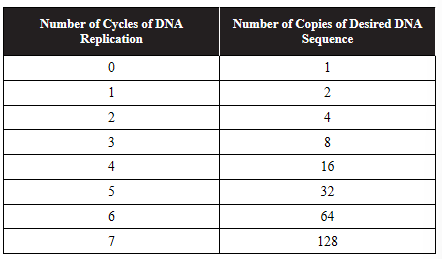
(a) Explain why a primer is needed to direct DNA polymerase to copy the desired sequence.
(b) Construct a graph of the data from the table.
(c) Analyze the data, and state the mathematical relationship between the number of cycles of PCR and the number of copies of the desired DNA sequence generated.
(d) Predict the minimum number of cycles required to generate 1,000 copies of the desired DNA sequence. Justify your prediction.
Practice Questions
Multiple-Choice
1. What would be the minimum number of nucleotides required to code for a protein made of 12 amino acids?
(A) 6
(B) 12
(C) 36
(D) 48
2. A scientist inserts a eukaryotic gene directly into a bacteria’s genome. However, the protein produced by the bacteria from the eukaryotic gene does not have the same amino acid sequence as the protein produced from the gene in eukaryotic cells. Which of the following best explains this?
(A) Prokaryotes and eukaryotes do not use the same genetic code, so the bacteria cannot decode the eukaryotic gene.
(B) Eukaryotic genes contain introns, which must be removed before translation; prokaryotes cannot remove introns.
(C) Prokaryotes do not have a nucleus and cannot perform transcription.
(D) Prokaryotes do not have rough endoplasmic reticulum, so they cannot translate eukaryotic genes.
3. Which of the following correctly represents the mRNA sequence that would be transcribed from the DNA sequence 3′ ACC GGT AAG TTC 5′?
(A) 3′ TGG CCA TTC AAG 5′
(B) 3′ UGG CCA UUC AAG 5′
(C) 5′ TGG CCA TTC AAG 3′
(D) 5′ UGG CCA UUC AAG 3′
4. What is the amino acid sequence that would be translated from the following gene? (Use Figure 16.4 to answer this question.)
3′ AAT CGT TTC AAT CAA 5′
(A) Asn-Arg-Phe-Asn-Gln
(B) Leu-Ala-Lys-Leu-Val
(C) Phe-Arg-Asn-Phe-Gln
(D) Gly-Ala-Lys-Gly-Val
5. Which of the following processes is most similar in prokaryotes and eukaryotes?
(A) alternative splicing of exons
(B) addition of a 3′ poly-A tail to mRNA
(C) addition of a 5′ GTP cap to mRNA
(D) transcription by RNA polymerase
6. The following figure depicts steps in the flow of genetic information in a eukaryotic cell.
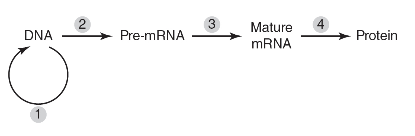
During which step will spliceosomes remove introns?
(A) 1
(B) 2
(C) 3
(D) 4
7. The antibiotic tetracycline inhibits bacterial growth by blocking the binding of tRNAs to the ribosome. Which of the following processes is most directly affected by tetracycline?
(A) transcription
(B) exon splicing
(C) initiation of translation
(D) export of proteins from the cell
8. Why do retroviruses have a high mutation rate?
(A) RNA polymerase has a high error rate when reading viral genomes.
(B) DNA polymerase has a high error rate when reading viral genomes.
(C) Viruses use a different genetic code than prokaryotes and eukaryotes use.
(D) Reverse transcriptase has a high error rate.
9. Humans can generate over 1012 different antibody proteins, but humans have fewer than 25,000 genes. Which of the following best explains how this is possible?
(A) Humans acquire new antibody genes when they are infected with pathogens.
(B) Alternative splicing of exons can generate many different transcripts from the same gene.
(C) The error rate in RNA polymerase generates new transcripts for antibody proteins.
(D) Golgi bodies modify RNA transcripts to create new combinations.
10. Which of the following catalyzes the formation of peptide bonds during translation?
(A) RNA polymerase
(B) mRNA
(C) rRNA
(D) tRNA
Short Free-Response
11. Ehlers-Danlos syndrome (EDS) type IV is a result of a mutation in the COL3A1 gene, which results in the deletion of one of the exons in the procollagen transcript.
(a) Describe the location in the cell where the splicing of exons occurs.
(b) Explain the difference between exons and introns.
(c) Predict the effect of the EDS mutation on the structure of the procollagen protein.
(d) Justify your prediction from part (c).
12. A mutation results in the deletion of the TATA box in the promoter of a eukaryotic gene.
(a) Describe the function of the promoter in gene expression.
(b) Explain why promoter sequences are highly conserved in living organisms.
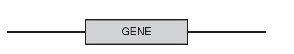
(c) The figure that follows shows the gene and the DNA surrounding it. Draw an “X” on the most likely location of the promoter of the gene.
(d) Explain how the deletion of the TATA box in the promoter would most likely affect the levels of protein produced by the gene.
Long Free-Response
13. The levels of mRNA and protein produced by three different genes (PDHA1, MBP, and INS) were measured in different tissues of the human body. The levels of mRNA and protein expression from these genes were compared to mRNA and protein expression from the gene POLR2A (RNA polymerase II subunit A, which is expressed in all human tissues). The results are shown in the table. “0” indicates that mRNA or protein were not detected in that tissue, and “+,” “++,” and “+++” indicate relatively low, medium, or high levels of mRNA or protein detected, respectively.
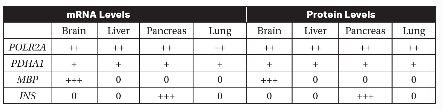
(a) Identify which gene was used as a control in this experiment.
(b) Explain why the gene you selected in part (a) would be an appropriate control for this experiment.
(c) Based on the data in the table, determine which of the genes (PDHA1, MBP, or INS) is most likely involved in glycolysis.
(d) INS is the insulin gene. Insulin is a hormone that is released when blood sugar levels are high. Predict the levels of INS mRNA and protein that would be found in cells in the pancreas when blood sugar levels are low. Justify your prediction.
Practice Questions
Multiple-Choice
1. A repressible operon in bacteria codes for the genes required to produce the amino acid serine. Serine functions as a corepressor for the operon. If serine is present in the bacteria’s environment, which of the following is most likely?
(A) increased digestion of serine
(B) increase in the levels of serine
(C) increased production of the repressor protein
(D) increased binding of the repressor protein to the operator
2. The arabinose operon is an inducible operon that codes for the genes required to digest the sugar arabinose. Arabinose functions as an inducer molecule for the operon. If arabinose is present in the bacteria’s environment, which of the following is most likely?
(A) increased digestion of arabinose
(B) increase in the levels of arabinose
(C) increased production of the repressor protein
(D) increased binding of the repressor protein to the operator
3. Which sequences in bacterial operons are noncoding sequences?
(A) operators and promoters only
(B) operators, promoters, and genes for regulatory proteins
(C) operators, promoters, and genes for structural proteins
(D) promoters, genes for regulatory proteins, and genes for structural proteins
4. Which of the following is a key difference between inducible operons and repressible operons?
(A) Inducible operons have promoter sequences, and repressible operons do not have promoter sequences.
(B) Inducible operons have operator sequences, and repressible operons do not have operator sequences.
(C) The repressor in an inducible operon can bind to the operator sequence on its own, but the repressor in a repressible operon requires a corepressor in order to bind to the operator.
(D) In inducible operons, RNA polymerase requires the presence of a corepressor in order to bind to the promoter; in repressible operons, RNA polymerase does not require a corepressor.
5. Under which of the following conditions will transcription of the lac operon be at its highest level?
(A) low glucose, low lactose
(B) low glucose, high lactose
(C) high glucose, low lactose
(D) high glucose, high lactose
6. Adding a methyl group to a DNA nucleotide is a type of _____________ and will make a DNA sequence much less likely to be transcribed.
(A) mutation
(B) epigenetic change
(C) aneuploidy
(D) transposition
7. Which of the following are proteins in eukaryotes that bind to regulatory switches and upregulate gene expression?
(A) activators
(B) repressors
(C) transcription factors
(D) mediators
8. Not every change in the DNA sequence results in a change in the amino acid sequence of a protein. Which of the following explains this?
(A) Each organism lives in a different environment, which changes the expression of its genes.
(B) The genetic code is redundant, and more than one codon codes for most amino acids.
(C) The proofreading function of ribosomes corrects changes in the DNA sequence.
(D) Differential gene expression adapts to these changes in the DNA sequence.
9. Which of the following does not affect the phenotype of an organism?
(A) the genes that are expressed
(B) the level at which genes are expressed
(C) the timing of gene expression
(D) the number of genes in the organism
10. Which of the following methods of horizontal transmission of genetic material would be most likely to lead to new variants of the COVID-19 virus?
(A) transformation
(B) transduction
(C) transposition
(D) conjugation
Short Free-Response
11. One way that organisms respond to changing environmental conditions is through the regulation of gene expression.
(a) Describe the function of operators and promoters in prokaryotes.
(b) Explain how the operator and repressor interact to control the expression of the inducible lac operon.
(c) Bacteria are placed in an environment that has low levels of glucose and high levels of lactose. Predict the level of expression of the lac operon in this environment.
(d) Justify your prediction from part (c).
12. A student conducts an experiment in an effort to determine whether a specific bacterial operon is inducible or repressible. The level of transcription of the operon was measured after the addition of different molecules to the bacteria’s environment. Data are shown in the table.
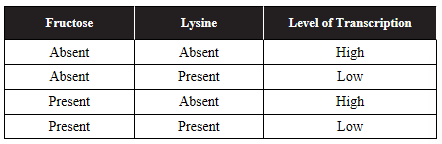
(a) Describe what, if any, effect levels of fructose have on the level of transcription of the operon.
(b) Describe what, if any, effect levels of lysine have on the level of transcription of the operon.
(c) Make a claim about whether this operon is more likely inducible or repressible.
(d) Justify your claim from part (c) using evidence from the experiment and your knowledge of inducible and repressible operons.
Long Free-Response
13. The human THY-1 gene (Thy-1 cell surface antigen) codes for a cell surface glycoprotein that is thought to function as a tumor suppressor in certain types of cancer. In order to find possible locations of enhancer sequences for the THY-1 gene, scientists performed a series of DNA deletion experiments. In the experiments, different deletions were made in areas that were suspected to function as possible enhancer sequences, and the levels of THY-1 transcription were measured after each deletion. The following diagram shows the sequences deleted. A, B, C, D, and E represent different DNA sequences. 1, 2, and 3 represent suspected enhancer sequences. P is the promoter of the THY-1 gene. An X represents the deletion of that portion of the DNA sequence.
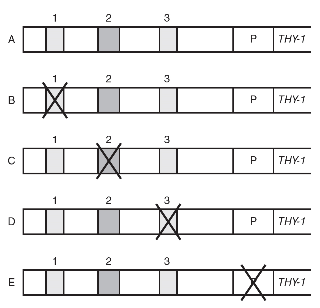
The levels of THY-1 transcription in each part of the experiment are shown in the graph.
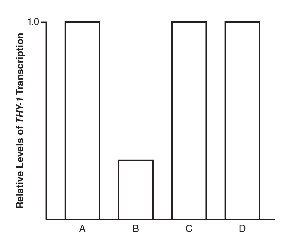
(a) Explain the function of enhancer sequences in eukaryotic gene expression.
(b) Identify which DNA sequence (A, B, C, D, or E) was the control in this experiment.
(c) Which sequence (1, 2, or 3) most likely functions as an enhancer sequence? Support your claim using evidence from the data.
(d) Predict the effects of the deletion of P, as shown in sequence E, and what the relative level of THY-1 transcription would be for E. Justify your prediction with evidence from the data.
Practice Questions
Multiple-Choice
1. Which of the following is used to cut DNA at specific base pair sequences?
(A) DNA ligase
(B) gel electrophoresis
(C) polymerase chain reaction
(D) restriction enzymes
2. A forensic scientist recovers a very small amount of evidence at a crime scene. The scientist would like to amplify the number of copies of DNA in the evidence sample. Which of the following techniques should the scientist use?
(A) bacterial transformation
(B) CRISPR-Cas9
(C) gel electrophoresis
(D) polymerase chain reaction
3. A forensic scientist cuts DNA from a crime scene and DNA from a suspect with the same enzyme. Which tool should the scientist use to separate the DNA fragments according to their size?
(A) bacterial transformation
(B) CRISPR-Cas9
(C) gel electrophoresis
(D) polymerase chain reaction
Questions 4 and 5
A scientist at a pharmaceutical company wants to create bacteria that will synthesize human growth hormone.
4. What should the scientist use to add the DNA code for human growth hormone to a plasmid?
(A) bacterial transformation
(B) DNA ligase
(C) gel electrophoresis
(D) polymerase chain reaction
5. The scientist has successfully created a plasmid that contains the DNA code for the human growth hormone gene. Which technique should the scientist use to insert the plasmid into a cell?
(A) bacterial transformation
(B) CRISPR-Cas9
(C) gel electrophoresis
(D) polymerase chain reaction
Questions 6–8
Huntington’s disease is caused by a short tandem CAG repeat in the HTT gene. Individuals with fewer than 35 CAG repeats in the HTT gene do not develop Huntington’s disease. Individuals with 40 or more CAG repeats will develop Huntington’s disease.
6. Which of the following tools would be most useful in amplifying the number of copies of the HTT gene so that more DNA would be available for analysis?
(A) CRISPR-Cas9
(B) gel electrophoresis
(C) polymerase chain reaction
(D) restriction enzymes
7. Which tool would be most useful for estimating the size of the HTT gene isolated?
(A) DNA ligase
(B) gel electrophoresis
(C) polymerase chain reaction
(D) restriction enzymes
8. A transgenic mouse has the HTT gene. A scientist wants to investigate whether inserting a stop codon into the HTT gene might prevent the mouse from developing the disease. Which of the following tools would be most useful in inserting a stop codon into the HTT gene?
(A) CRISPR-Cas9
(B) gel electrophoresis
(C) polymerase chain reaction
(D) restriction enzymes
_________________________________________________________________________
(A) the nitrogen atoms in the nitrogenous bases
(B) the hydrogen bonds that form between the bases in the two strands of the double helix
(C) the charges on the oxygen atoms in the deoxyribose sugars
(D) the charges on the phosphates in the backbone of the helix
10. Why is a heat-stable DNA polymerase required for the polymerase chain reaction (PCR)?
(A) Heat-stable DNA polymerases add DNA nucleotides at a faster rate than other DNA polymerases.
(B) The high temperatures that are required for each cycle of PCR to separate the DNA strands would denature DNA polymerases that are not heat stable.
(C) Heat-stable DNA polymerases anneal more readily to the PCR primers.
(D) Heat-stable DNA polymerases are more accurate than other DNA polymerases.
Short Free-Response
11. The pAMP-pKAN plasmid contains both the ampicillin resistance gene and a kanamycin resistance gene.
(a) Describe a method for inserting this plasmid into a bacterial cell.
(b) Explain how one could detect which bacterial cells had successfully absorbed the pAMP-pKAN plasmid.
(c) A restriction enzyme makes a cut in the kanamycin resistance gene. Predict the result if a bacterium absorbs this piece of DNA that has been cut by the restriction enzyme.
(d) Justify your prediction from part (c).
12. The comet assay technique is one way of measuring the relative amount of DNA damage in a cell. The nucleus of a cell is placed on a slide coated with an agarose gel. An electric current is applied to the gel, and smaller pieces of DNA will move farther away from the location where the nucleus was placed. These smaller pieces of DNA form the “tail,” and the nucleus forms the “head” of the “comet.” Longer tails indicate greater amounts of DNA damage.
(a) Explain why DNA fragments will move through an agarose gel when an electric current is applied.
(b) List and describe two things that could damage DNA and result in longer tails on the comet assay.
(c) Two cells are treated with a known mutagen of DNA. Cell A has two functional p53 alleles (p53 is involved in DNA repair). Cell B has one functional p53 allele, and its second p53 allele is nonfunctional. On the two templates provided, construct a visual representation of the comet assay results for these two cells.
(d) Explain the visual representations you created on the two templates in part (c).
Long Free-Response
13. A student conducts a bacterial transformation experiment using a strain of E. coli that is not resistant to antibiotics. The student also uses a plasmid (pAMP-pTET) that carries genes for both ampicillin and tetracycline resistance. The four different plates used, and the results of this experiment, are shown in the following figure.
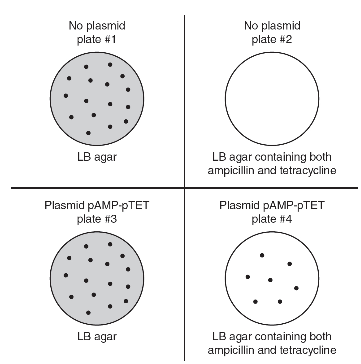
(a) Describe the two enzymes that would be used to create a recombinant plasmid that contained both the ampicillin resistance gene and the tetracycline resistance gene.
(b) Identify the plate that acts as a control to show that the untransformed bacteria were not originally resistant to ampicillin and tetracycline.
(c) Analyze the data, and determine whether the relative numbers of bacteria colonies (seen on agar plate #3 and agar plate #4) are as expected if the bacterial transformation procedure was successful.
(d) Bacteria are removed from plate #4 and are allowed to grow in nutrient medium. Bacteria that contain the F+ (fertility) plasmid can undergo the process of conjugation and transfer some of their DNA to other bacterial cells. Predict the result if bacteria (that were resistant to streptomycin and contained the F+ plasmid) were used to transform the bacteria from plate #4. Justify your prediction.




